17.6. Storage Subsystem¶
17.6.1. Use Cases¶
UC01: {Repository operators, researchers} need to upload large, or many files
Priority: 2 (blocked by UC04)
High volume network transfer, e.g., over Globus or other non-http protocols)
Bootstrap new MN install (e.g., ADC) with, say, 5TB of data over many files w/o paying the network transfer penalty associated with MNStorage.create
MN operators (e.g., ADC) loading large submissions. Currently might take hours/days. We might call this “out of band data loading”
Have alternative options for users that might have slow internet speeds (ability to resume where they might have left off)
Arctic Data Center, ESS-DIVE, PDG, DataONE
UC02: {Repository operators, researchers} want to create/update file system based data uploads
Upload a single file (data or metadata) to the file system
Upload a whole package with all of its metadata and data files in a package hierarchy
triggers creation of system metadata?
“zip-based” upload is start of support for create/update transactions and rollback
ESS-DIVE, rdataone, …
UC03: {Researchers} needs a hierarchical file system read-only view of packages
e.g., run analysis against files in package hierarchy
view should be read-only
need to enforce access control rules (e.g., only show readable files and packages)
UC04: {MN System tools} need direct access to metadata/data
Priority: 1 (blocks others)
priviledged tools that can access all data
only available to ‘admin’ users/tools
MetaDIG quality engine needs direct, efficient access to metadata/data
would enable metadata/data congruence checks
would enable advanced data checks
Need access to files from distributed clusters like Kubernetes
DataONE LOD service needs read-only access to metadata (possibly data). Currently gets it all over HTTP
Efficient data previews in the UI or other client tools
ESS-DIVE file checks of data (e.g., for virus checking)
Indexer needs access to on-disk metadata files and resource maps and system-metadata
Currently give file path to bytes for science metadata and resource maps,
system metadata comes from HZ, needs to be accessible from the filesystem too
Need access to files from distributed clusters like Kubernetes
System metadata needs to be stored on the filesystem
Service to create checksums for all data objects (i.e. Carl Bottinger)
a suite of checksums could be calculated and published to a registry
Metrics Service access to the metadata to parse citations,…
ADC, ESS_DIVE, DataONE,
UC05: {Researchers} create and trigger data quality assessment workflows
for Arctic Data Center, with user contributed data checking workflows
requires access to files on disk, arranged in hierarchical packages as the user “sees” it
UC06a: Downloading large packages via BagIt over HTTP
Need ability to download large packages in the face of network errors - HTTP Byte-range support
Fix our Bagit download implementation
The ability to download the files based on how on the hierarchical file system (eg. I want to download all the files from the same folder)
UC06: Downloading large packages via alternate protocols
Priority: 3
Bulk download is too difficult currently for people and I can imagine us being able to set up a permanent or temporary FTP or something to give durable access
Subcase 1: download all of Kate Stafford’s files
Requires a large bagit beyond our current capabilities
Subcase 2: mark and download a subset of the files in a package hierarchy
Subcase 3: Support OPeNDAP/THREDDS access
Globus support
Being able to download a specific group of data files (eg. all the data related to a sample)
UC07: {Researchers} want to remotely mount a filesystem view of their {package, collection, repository, DataONE}
Same as ONEDrive use cases from DataONE Phase I
UC08: {Researchers} want to upload their data packages via Google Drive/Box/Dropbox/OneDrive/GitHub
Fill in metadata somewhere, tag a release, get’s uploaded automatically
science metadata gets added on/layered on top of files obtained from shared drives, etc.
UC09: {Contributors} want to do batch updates to {large} number of existing files
Need a batch API for things like access control updates e.g. send a single API request for various metadata about a collection of objects (rather than sending one call for each type of metadata for each object)
https://hpad.dataone.org/fJtGygXLRH-pUGG4XnPDWw#
UC10: {Contributors} want to be able to upload and view files in folder structures
For both small and large packages
Involves how we rearchitect packages as large collections of files
ESS-DIVE use cases
some files are too large to upload to DataONE
links are made to the data
essdive defined access types
tier 1
in metacat, replicated
tier 2
not in metacat
tier 3
offline, private
requires request, manual access
direct access to data from Jupyter notebook
user creates a list of files to download, creating a collection like a shopping cart, then requests download
maybe a BagIt file is created
on upload
check that files are in required format
17.6.2. Requirements (Capabilities)¶
ability to upload very large files, efficiently (current Metacat limit is 5MB)
ability to upload many files (thousands), efficiently
provide high volume, non-HTTP access to Metacat data store
provide public or authenticated access depending on object
provide a hierarchical view of Metacat data store
files ingested/uploaded are integrated into DataONE as if they were uploaded via DataONE REST API
system metadata created
objects indexed in Solr
access via the storage API should have better performance than the DataONE REST API
for clients (e.g. MetaDIG enginee)
read access needed for all DataONE metadata
read access needed for potentially all DataONE data objects
more effiecient than downloading metadata for each run
read only access to hierarchical view of any data package
read only access to all DataONE system metadata
Batch updates in Metacat (https://hpad.dataone.org/fJtGygXLRH-pUGG4XnPDWw#)
17.6.3. Design diagrams¶
17.6.3.1. Architecture¶
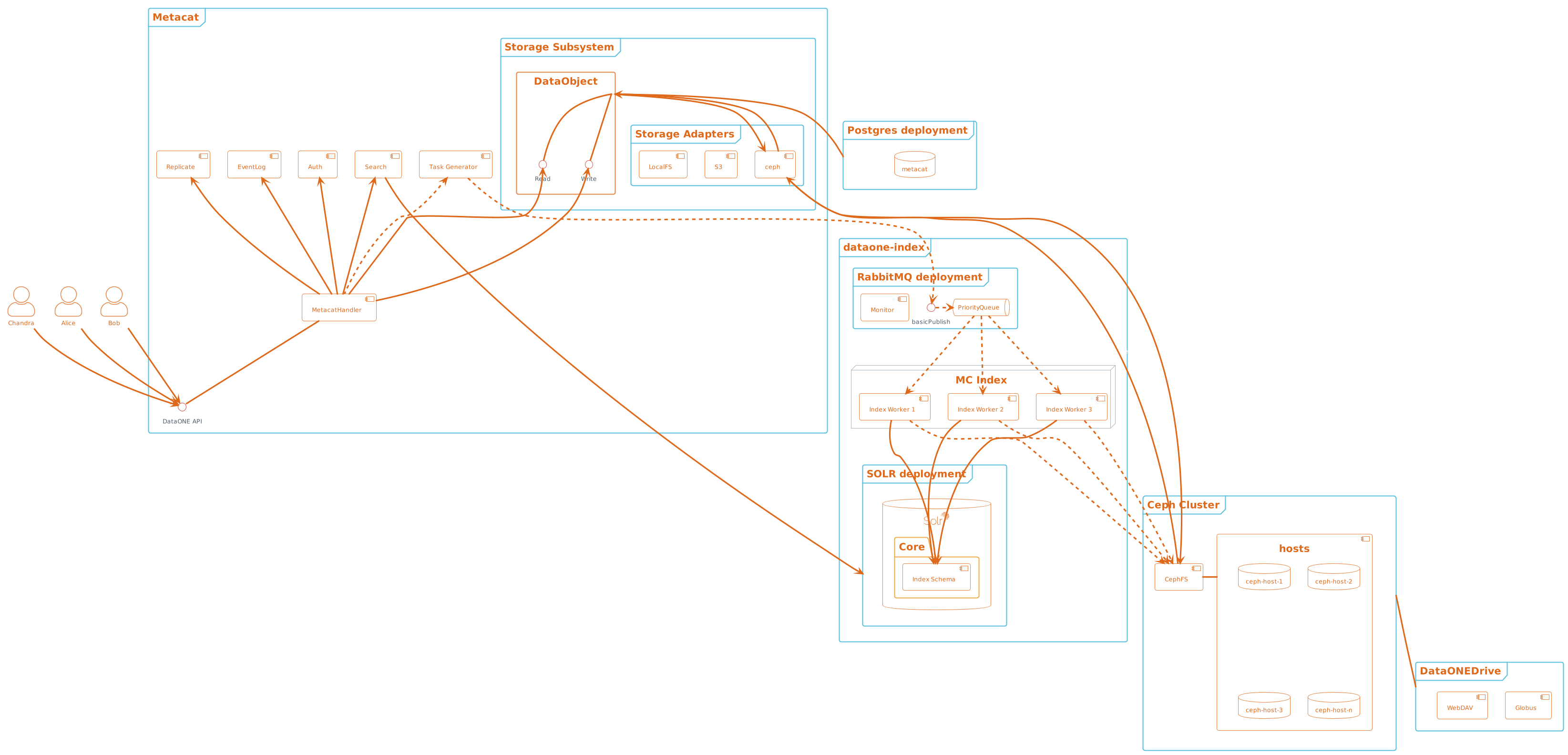
Figure 1. Metacat components overview, highlighting index task flow with dashed arrows.¶
Design choices include:
Storage interface
- Ceph FS
- Ceph Object Gateway
S3 API (see https://docs.ceph.com/en/latest/radosgw/s3/)
Swift API
Data and metadata access layer
- What level should we plan for access by:
System processes
User access processes
17.6.3.2. Physical File Layout¶
For physical file storage and layout, our goal is to provide a consistent directory structure that will be robust against naming issues such as illegal characters, and that allows us to access both system metadata and the file contents knowing only the PID for an object. This approach focuses on using the hash identifier of an authority-based identifier such as a PID or SID for naming objects (rather than the content identifier). The possibility that duplicate objects may be stored is accepted in favour of reducing the complexity and overhead involved in the object deduplication process.
The alternative approach of using the raw bytes of each object (content identifier) is discussed in: appendix/storage-subsystem-cid-file-layout.rst
Raw File Storage: The raw bytes of each object (data, metadata, or resource map) are saved in a file that is named using the hash of an authority-based identifier (PID) for that set of bytes. The resulting identifier (checksum value) is then used to name the file. Note, in this approach, a file can be uploaded multiple times under different checksums (PID hashes).
Checksum algorithm and encoding
We have multiple hash algorithms to choose from, and each has multiple ways of encoding the binary hash into a string representation. We will choose the simplest, most common configuration which is to use a SHA-256 hash algorithm, with the binary digest converted to a string value using base64 encoding. That makes each hash value 64 characters long (representing the 256 bit binary value). For example, here is a base64-encoded SHA-256 value:
4d198171eef969d553d4c9537b1811a7b078f9a3804fc978a761bc014c05972c
While we chose this common combination, we could also have chosen other hash algorithms (e.g., SHA-1, SHA3-256, blake2b-512) and alternate string encodings (e.g., base58, Multihash (https://multiformats.io/multihash/)). Multihash may be a valuable approach in to future-proof the storage system, because it enables us of multiple checksum algorithms.
Folder layout
To reduce the number of files in a given directory, we use the first several characters of the hash to create a directory hierarchy and divide the files up to make the tree simpler to explore and less likely to exceed operating system limits on files. We store all objects in an objects directory, with three levels of ‘depth’ and a ‘width’ of 2 digits (https://github.com/DataONEorg/hashstore/issues/3). Because each digit in the hash can contain 16 values, the directory structure can contain 16,777,216 subdirectories (256^3). An example file layout for three objects would be:
/var/metacat/objects
├── 7f
│ └── 5c
│ └── c1
│ └── 8f0b04e812a3b4c8f686ce34e6fec558804bf61e54b176742a7f6368d6
├── a8
│ └── 24
│ └── 19
│ └── 25740d5dcd719596639e780e0a090c9d55a5d0372b0eaf55ed711d4edf
└── 0d
└── 55
└── 5e
└── d77052d7e166017f779cbc193357c3a5006ee8b8457230bcf7abcef65e
Note how the full hash value is obtained by appending the directory names with the file name (e.g., 7f5cc18f0b04e812a3b4c8f686ce34e6fec558804bf61e54b176742a7f6368d6 for the first object).
Storing metadata With this layout, knowing the hash value for a PID allows us to retrieve it. But it does not provide a mechanism to store metadata about the object, other system metadata for the object, or extended metadata that we might want to include. So, in addition to data objects, the system supports storage for metadata documents that are associated with any desired data object. These metadata files are stored in the metadata directory parallel to objects, and structured analogously. Additional context can be found here (https://github.com/DataONEorg/hashstore/issues/35).
For example, given the PID ‘jtao.1700.1’ and formatId ‘http://ns.dataone.org/service/types/v2.0’, one can calculate the location of its metadata document using:
$ echo -n "jtao.1700.1http://ns.dataone.org/service/types/v2.0" | shasum -a 256
ddf07952ef28efc099d10d8b682480f7d2da60015f5d8873b6e1ea75b4baf689
So, the system metadata file would be stored in the metadata directory, with the address ‘/dd/f0/79/52ef28efc099d10d8b682480f7d2da60015f5d8873b6e1ea75b4baf689’ using the file format described above. Extending our diagram from above, we now see the three hashes that represent data files, along with three that represent system metadata files named with the hash of the PID+formatId:
/var/metacat
├── objects
│ ├── 7f
│ │ └── 5c
│ │ └── c1
│ │ └── 555ed77052d7e166017f779cbc193357c3a5006ee8b8457230bcf7abcef65e
│ ├── a8
│ │ └── 24
│ │ └── 19
│ │ └── 25740d5dcd719596639e780e0a090c9d55a5d0372b0eaf55ed711d4edf
│ └── 0d
│ └── 55
│ └── 5e
│ └── d77052d7e166017f779cbc193357c3a5006ee8b8457230bcf7abcef65e
└── metadata
├── 9a
│ └── 2e
│ └── 08
│ └── c666b728e6cbd04d247b9e556df3de5b2ca49f7c5a24868eb27cddbff2
├── dd
│ └── f0
│ └── 79
│ └── 52ef28efc099d10d8b682480f7d2da60015f5d8873b6e1ea75b4baf689
└── 32
└── 3e
└── 07
└── 99524cec4c7e14d31289cefd884b563b5c052f154a066de5ec1e477da7
PID-based access: Given a PID and a formatId, we can discover and access both the system metadata for an object and the bytes of the object itself without any further store of information (if no formatId is supplied, we will default to the agreed upon formatId for Hashstore (i.e. “http://ns.dataone.org/service/types/v2.0”).
The procedure for this is as follows:
Given the PID, calculate the SHA-256 hash, and base64-encode it to find pid hash of the data object.
Use the SHA-256 hash of the PID + formatId to locate and find the metadata object from the metadata tree
With the pid hash, open and read the data from the objects tree. With the hash of the PID + formatId, open and read data from the metadata tree.
Other metadata types: While we currently only have a need to access system metadata for each object, in the future we envision potentially including other metadata files that can be used for describing individual data objects. This might include package relationships and other annotations that we wish to include for each data file.
To pre-emptively accommodate this need, we have revised HashStore to store ‘metadata’, not only ‘sysmeta’. All metadata files will be stored in the metadata directory, with the permanent address being the SHA-256 hash of the pid+formatId and broken up into directory depths and widths as defined by a configuration file ‘hashstore.yaml’. This configuration file is written by HashStore upon successful verification that a HashStore does not exist.
17.6.3.3. Public API¶
While Metacat will primarily handle read/write operations, other services like MetaDig and DataONE MNs may interact with the hashstore directly. Below are the public methods implemented in the Python implementation and Java implementation of HashStore. These are pending review and integration into Metacat.
The methods below will be included in the public API:
Method |
Args |
Return Type |
Notes |
|---|---|---|---|
store_object* |
pid, data, … |
hash_address (object_cid, …) |
Pending Review |
store_metadata |
pid, sysmeta, format_id |
metadata_cid |
Pending Review |
retrieve_object |
pid |
io.BufferedIOBase |
Pending Review |
retrieve_metadata |
pid, format_id |
string (metadata) |
Pending Review |
delete_object |
pid |
boolean |
Pending Review |
delete_metadata |
pid, format_id |
boolean |
Pending Review |
get_hex_digest |
pid, algorithm |
string (hex_digest) |
Pending Review |
store_object(pid, data, additional_algorithm, checksum, checksum_algorithm)
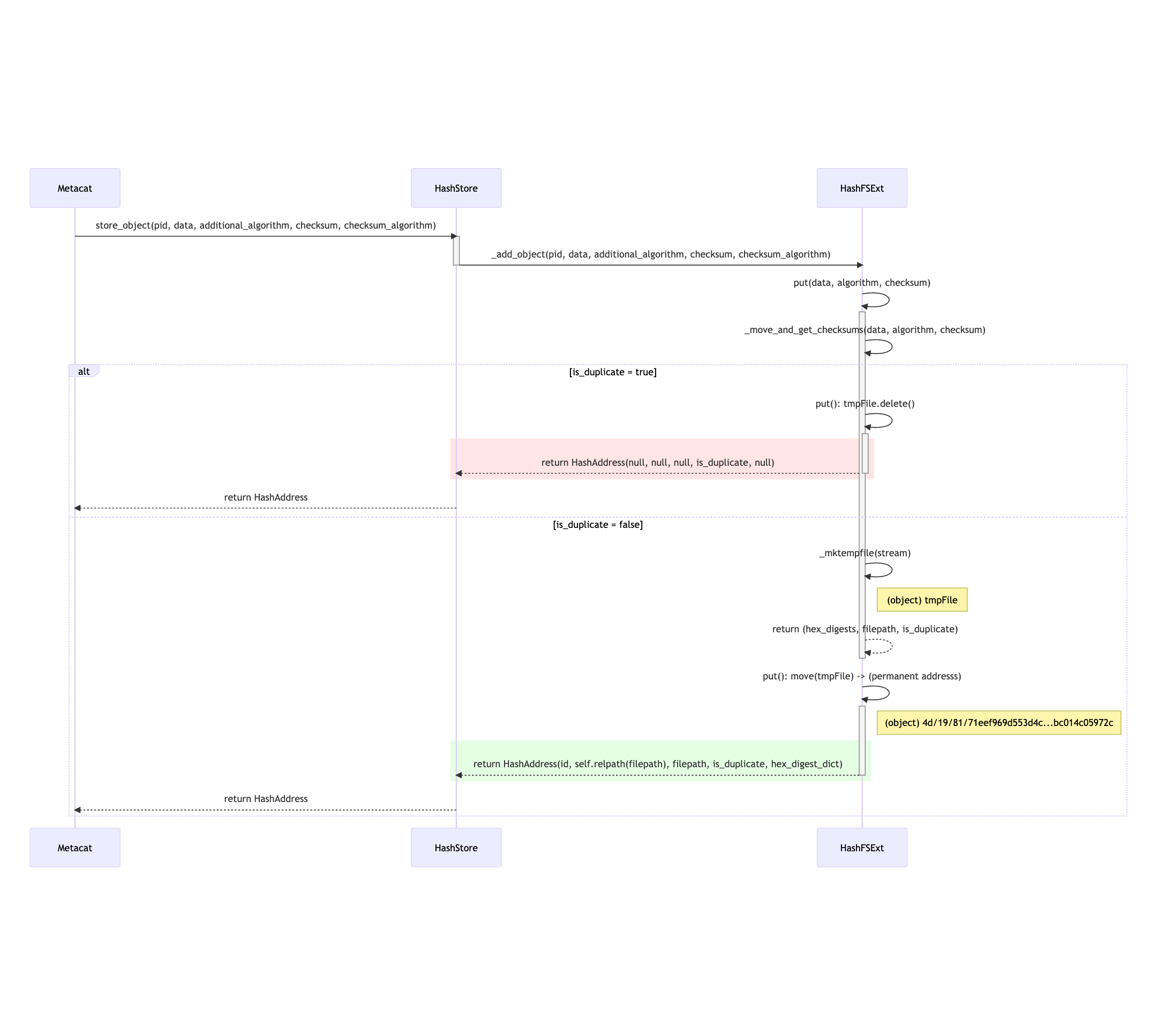
Figure 2. Mermaid diagram illustrating store_object flow¶
17.6.3.4. Annotations¶
To support a paging API/query service to parse large data packages, we are proposing that each member (subject) in a dataset package will have a RDF annotation file formatted in either N-Triples or JSON-LD. Each annotation file can contain multiple triples that describe the subject’s relationships with its respective objects. These files will exist alongside sysmeta documents in HashStore, and can be stored, retrieved and deleted with the same public API. In the diagram below, we illustrate this concept:
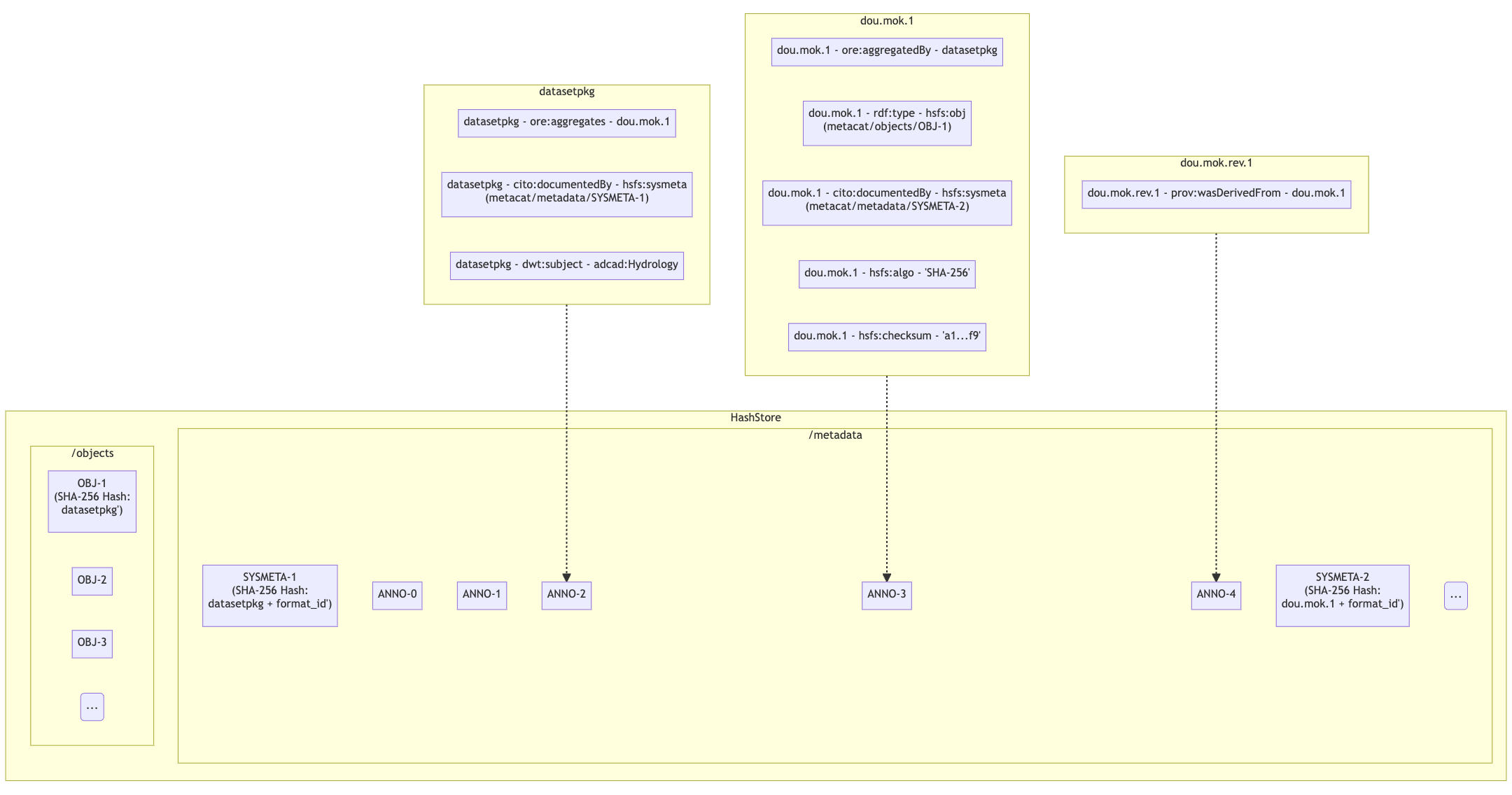
- Notes:
Each annotation file (prefixed by ANNO-) contains triples about a subject, and the dotted line ending with an arrow represents where the annotation file would exist in HashStore
Ex. ‘dou.mok.1’ has 5 triples, describing what package it belongs to, that it’s an object that can be found in HashStore, with a sysmeta document that can also be found in HashStore and a checksum and checksum algorithm.
To actually query the annotation graph, we will require an indexer that can not only index these annotation files, but also be able to merge what’s in additional ORE and EML documents to provide completeness to the graph. This index serves as the basis for the query service that MetacatUI/front-end would access. Users would not be accessing HashStore/Metacat directly, but this indexed version instead.
17.6.3.5. Virtual File Layout¶
In both of these cases, the main presentation of the directory layout would be via a virtual layout that uses human-readable names and a directory structure derived from the prov:atLocation metadata in our packages. Because a single file can be a member of multiple packages (both different versions of the same package, and totally independent packages), there is a 1:many mapping between files and packages. In addition, a file may be in different locations in these various packages of which it is a part. So, our ‘virtual’ view will be derived from the metadata for a package, and will enable us to browse through the contents of the package independently of its physical layout.
Within the virtual package display, the main data directory will be reflected at the root of the tree, with a hidden .metadata directroy containing all of the metadata files.
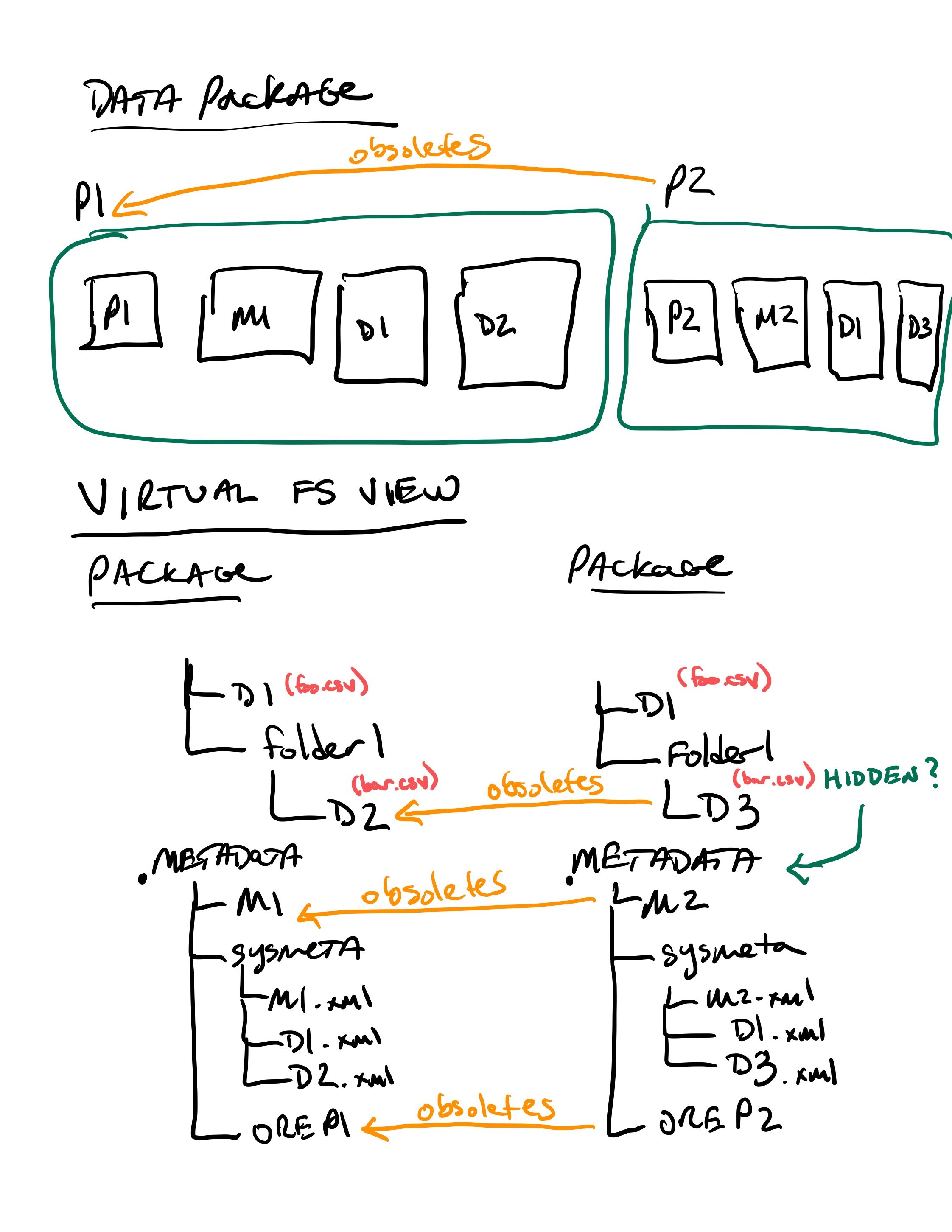
Virtual file layout.¶
This layout will be familiar to researchers, but differs somewhat from the BagIt format used for laying out data packages. In the BagIt approach, metadata files are stored at the root of the folder structure, and files are held in a data subdirectory. Ideally, we could hide the BagIt metadata manifests in a hidden directory and keep the main files at the root.
17.6.3.6. Filesystem Mounts¶
To support processing these files, we want to mount the data on various virtual machines and nodes in the Kubernetes cluster so that mutliple processors can seamlessly update and access the data. Thus our plan is to use a shared virtual filesystem. Reading from and writing to the shared virtual filesystem will result in reads and writes against the files in the physical layout using checksums.
17.6.3.7. Legacy MN Indexing¶
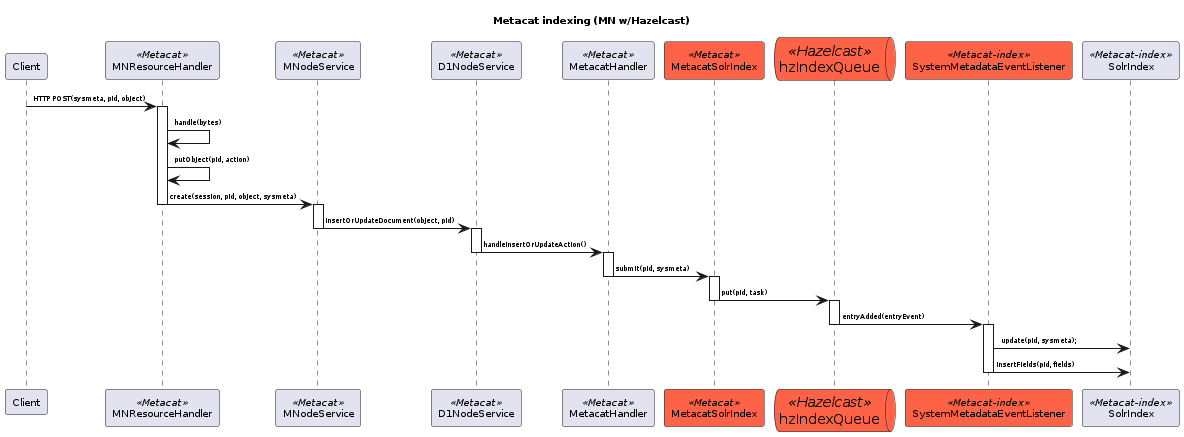
This sequence diagram shows the member node indexing message/data flow as it existed before the Metacat Storage/Indexing subsystem. The red boxes represent components that could be replaced by RabbitMQ messaging.¶
17.6.3.8. Legacy CN Indexing¶
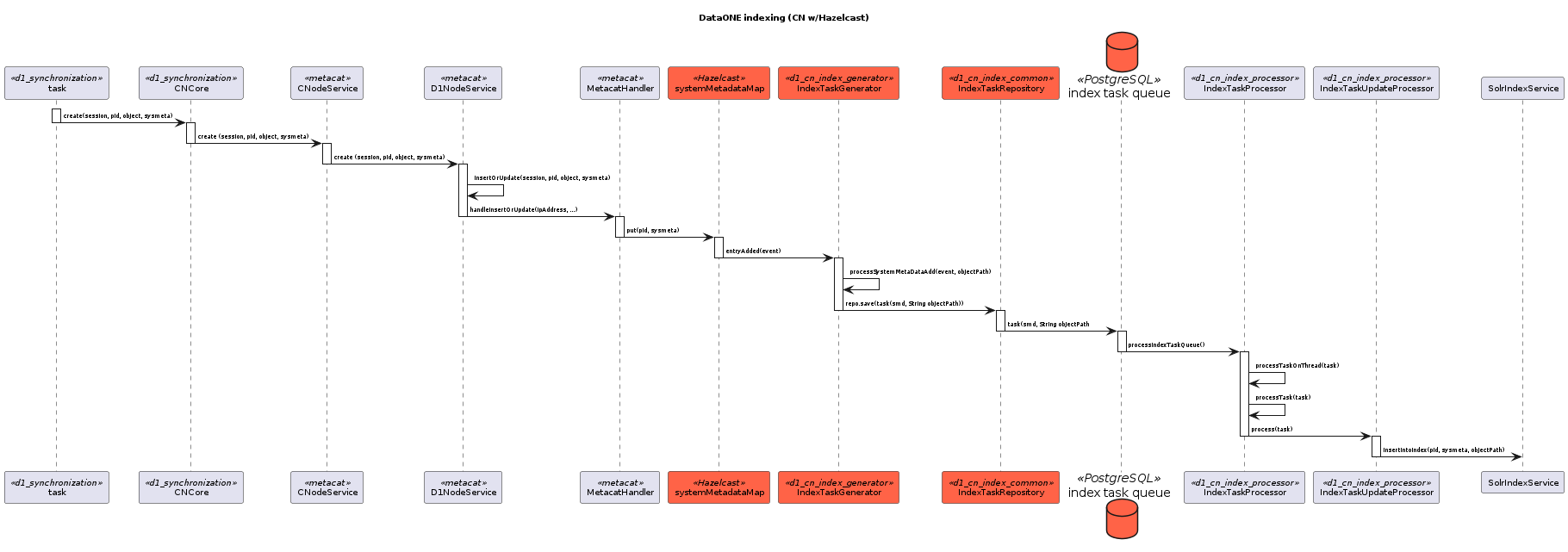
This sequence diagram shows the coordinating legacy indexing message/data flow as implemented before the Metacat Storage/Indexing subsystem. The red boxes represent compoenents that could be replaced by RabbitMQ messaging.¶
17.6.3.9. Proposed Indexing with RabbitMQ¶
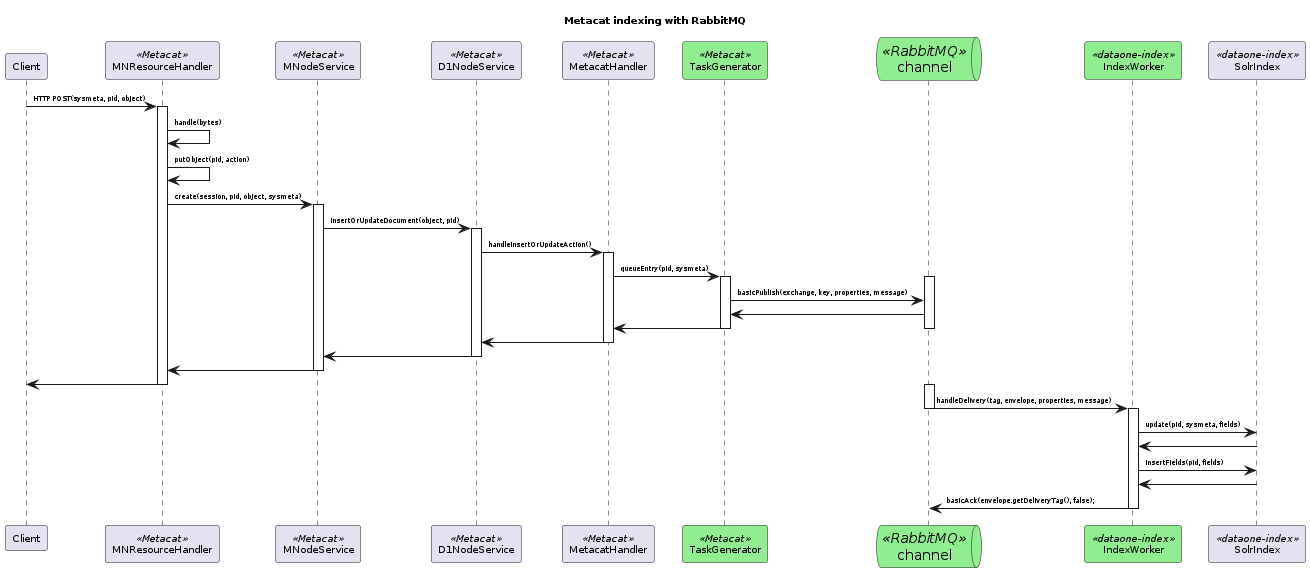
This sequence diagram shows indexing message/data flow as it could be refactored to use RabbitMQ. The green boxes represent compenents that would replace legacy indexing components in order to implement RabbitMQ messaging.¶
17.6.3.10. Proposed MN.create method¶
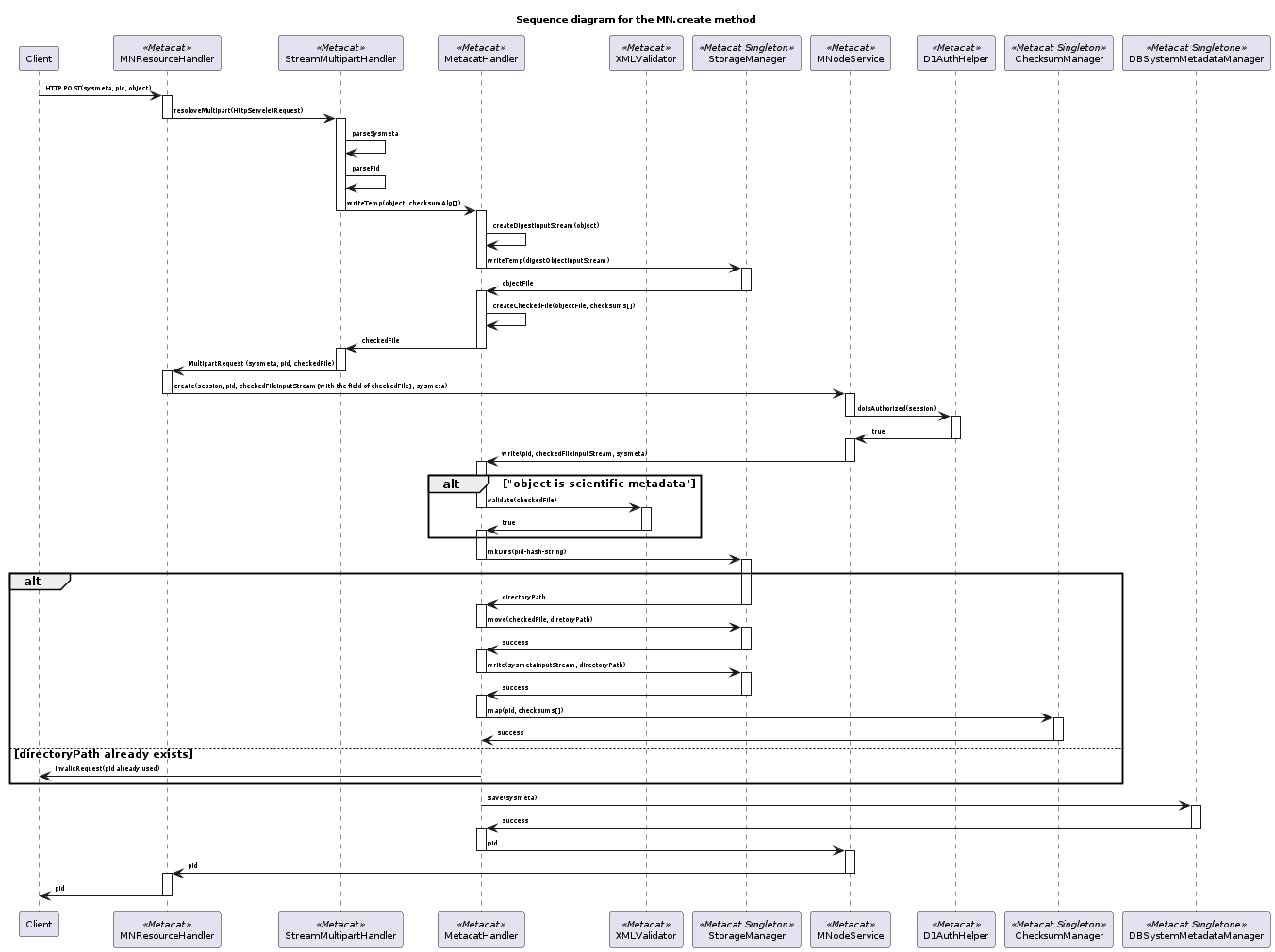
This sequence diagram shows data flow in the MN.create method.¶
17.6.3.11. CheckedFile and CheckedFileInputStream Class Diagram¶